Section 13 Partial Least Squares (PLS)
Integrate two datasets with PLS (Partial Least Squares analysis).
Unravel relationships between two numeric datasets acquired on the same matched individuals
- Pre-processing to ensure individuals of both datasets match (numerical description of combined datasets, Venn diagramm and upset plot)
- (Similarly to PCA) Graphical outputs: screeplot, projection of individuals, projection of variables on circle of correlation.
- Extract a new dataset based on PLS loadings or on selected variables.
The method used in ASTERICS comes from the mixOmics (Rohart et al. 2017) package.
For further information on PLS method:
13.1 Preprocessing

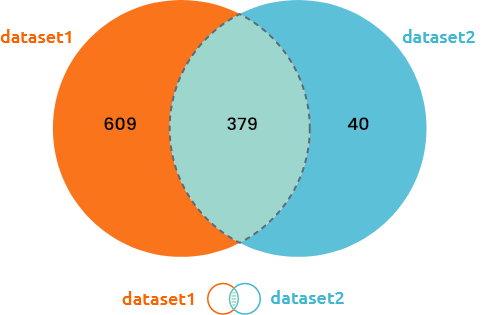
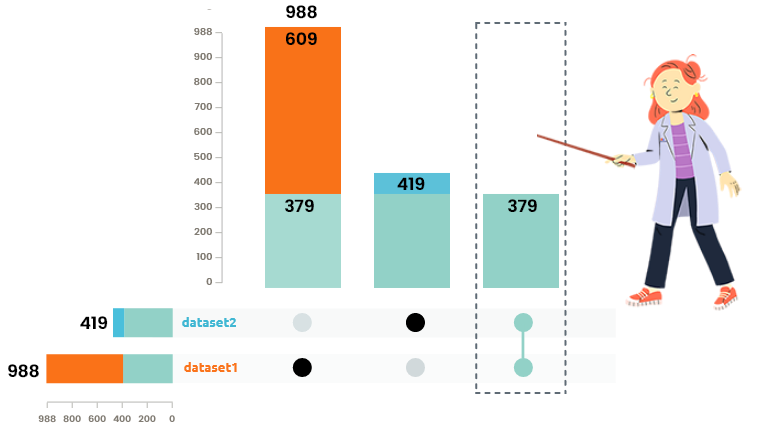
Venn diagram and upset plots are used to understand how many individuals are common / specific to each dataset. Only individuals common to all integrated datasets are used in the analysis.
13.2 Run PLS
How to set options?
Now that your datasets are ready to be used, click on the left panel to start PLS and wait for the computation (that can be a bit long).Important note: PLS is performed with raw values (data are not scaled to unit variance before they are processed). If you want to perform PLS on scaled data (for one or both datasets), go to the menu “Edit / Dataset edition” and scale your dataset(s) before PLS.

- in the “canonical” mode, the objective is to unravel the main relations between the two numeric datasets in a symmetric manner;
- in the “regression” mode, the objective is to explain at best one of the two datasets from the other.
In ASTERICS, the performed analysis is of the “canonical” mode unless one of the two datasets has only one or two variables (columns). In this case, PLS is automatically performed in “regression” mode with this dataset the one to be explained from the other.
Contrary to PCA, reproduced inertia is not an objective of PLS. These plots are given as mere information but can not be directly use to choose the number of PLS components.
13.3 Explore individuals

Similarly to PCA, the interpretation of PLS is done component (axis) by component, starting from the first which displays the main relationships between variables from the two datasets.
Combine the plot of individuals with colors (or shapes / sizes) giving information on other variables (e.g., variables of your design) to check if colors are organized with respect to components.
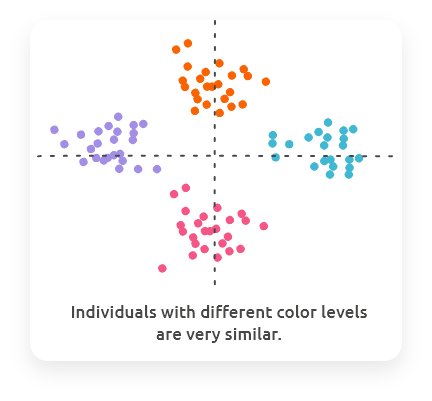
In regression mode (when one of the two datasets has only one or two variables), the variables to explain (the one in the dataset with one or two variables) are represented by the color and size of the dots in the plot of individuals if the user did not make another choice.

In regression mode (when one of the two datasets has only one or two variables), individuals are projected on the X-variate space only, whereas in canonical mode, they are projected on a consensual XY-variate space.
13.4 Explore variables
How to set options?
By choosing a correlation threshold, only variables with a correlation larger than this number are displayed on the plot (to make it easier to read).

Only variables well correlated with axes can be interpreted. Select a correlation threshold to display the most correlated variables.

You may display the variables from the
two datasets using “Separated” to improve the readability of the plots but be
careful that the interpretation has to be done with overlapping X- and Y-
representations.
In regression mode, on the contrary, only the X-representation is available.
13.5 Extract new data
How to set options?
Set options in left panel to generate a new dataset from the analysis:
with the criterion “Components, by number,” the first components (number to be specified by the user) will be extracted and used as a new dataset;
with the criterion “Components, by explained variance” (when available), the first components (number automatically set to reach the targeted percentage of explained variance or a targeted correlation ratio for categorical variables) will be extracted and used as a new dataset;
with the criterion “Variables,” the variables the most correlated with the first components will be selected and used as a new dataset.
When the dataset is extracted, you can use it in other analyses or check it in menu “My workspace.”
13.6 Default parameters
Used function and default parameters:
- If Y has 1 or 2 variables (columns):
- mixOmics::pls with options
ncomp = min(ncol(X), 20),mode = 'regression'andscale = FALSE.
- mixOmics::pls with options
- Otherwise:
- mixOmics::pls with options
ncomp = min(ncol(X), ncol(Y), 20),mode = 'canonical'andscale = FALSE.
- mixOmics::pls with options