Section 6 Missing values
Under this menu, you can handle the missing values in your datasets:
- Explore the distribution of missing values in your dataset
- Impute missing values with PCA, k-means or by zero
- OR remove individuals / variables with the largest proportions of missing values
6.1 Explore missing values
6.1.1 Heatmaps

Visually compare the two heatmaps (raw/organized):
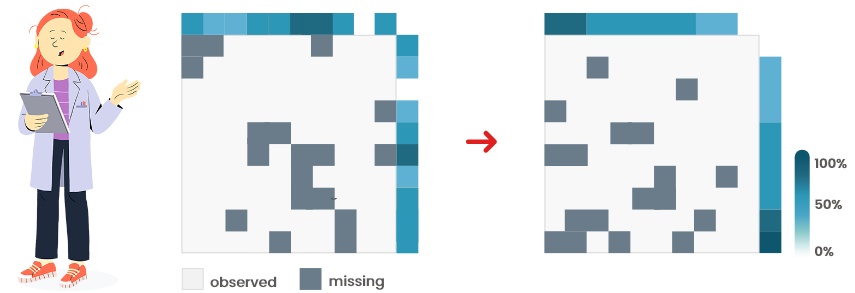
Figure 6.1: Heatmaps (without pattern of missingness)
They look alike (Figure 6.1): the values are probably missing completely at random.
Consider imputing missing values (if the proportion of missing is not too high,
smaller than 30% for instance).
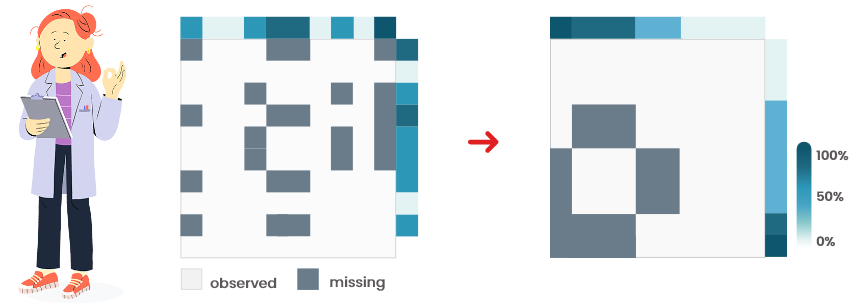
Figure 6.2: Heatmaps (with pattern of missingness)

They are very different (Figure 6.2): this indicates specific
patterns of missing values that might not be random.
Imputation or row/column removal must be performed with care because they might
induce biases in subsequent analyses.
6.1.2 Barplots

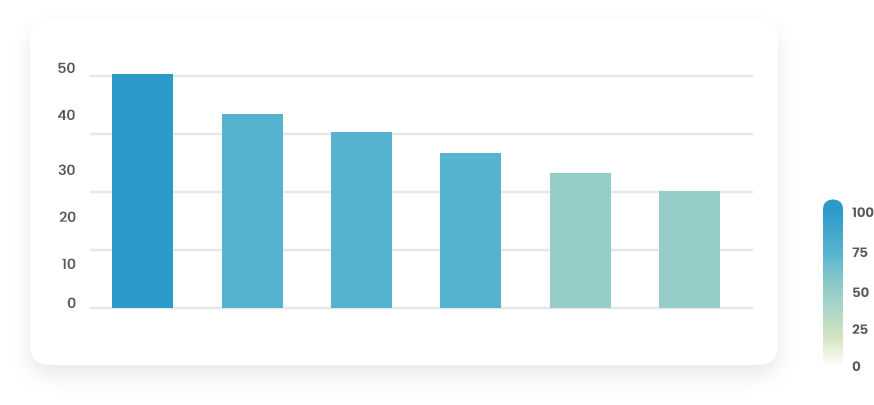
Figure 6.3: Barplot with regular decreasing shape
The barplot (Figure 6.3) has a regular decreasing shape with roughly equal steps: this indicates that values are probably missing completely at random. Consider imputing missing values (if the proportion of missing is not too high, smaller than 30% for instance).
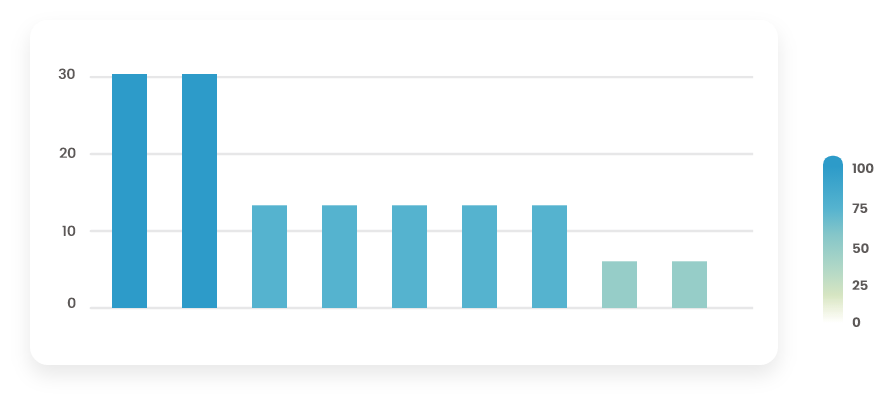
Figure 6.4: Barplot with steps

The (Figure 6.4) barplot has steps with various heights. Imputation or row/column removal must be performed with care because they might induce biases in subsequent analyses. Does the pattern make sense? Is it possible to link missingness patterns with a factor of the experimental design? If so, consider imputing by the levels of this factor.
6.2 Remove missing values
6.2.1 How to set options?
- “Direction” is used to choose to remove either entire individuals (rows) or entire variables (columns).
- The chosen percentage corresponds to the maximum acceptable missingness. For instance, if 30% is chosen, it means that all individuals or variables with more than 30% of missing values will be removed.

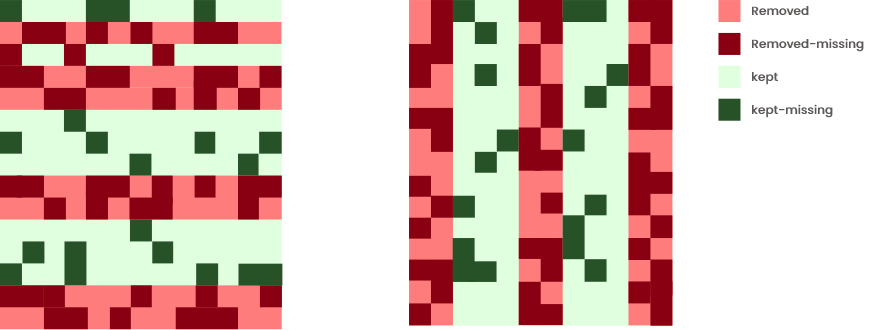
Here entire individuals (rows) or variables (columns) are removed (based on a maximum acceptable percentage of missingness). Be aware that, in all cases, this method induces strong biases when values are not missing at random. However, it is advised to use it if all missing values are concentrated in a few individuals and/or variables.
Check which method and percentage leads to the higher number of missing values removed (dark red), while maintaining a small number of non missing values removed (light red). Remaining missing values after removal of the rows/columns are in dark green.
When the resulting dataset is satisfactory, save it by clicking on
![]() .
.
6.3 Impute missing values
6.3.1 How to set options?
Imputation method can be chosen according to what you know on the missing values and subsequent analyses:
- imputation by zeros is dedicated to cases where missing values are due to a measurements below the detection threshold. It is a very basic > > approach to address this problem but certainly not the best;
- imputation by PCA is well designed when you want to use PCA, MFA, or PLS (for instance) afterwards because it best preserves the projection of your individuals on PC axes;
- imputation by k-nearest neighbors is based on the idea that two individuals that are similar on observed values also have similar values for unobserved variables. It best preserves the distances between individuals and is well adapted prior clustering.
The last two methods are only valid when data are missing at random.
In addition, for PCA and KNN you can choose to impute only certain types of variables (only numerical or only categorical variables). Setting this option to “Auto” imputes only variables of the most present type while “Mixed” imputes both numerical and categorical variables.
When the resulting dataset is satisfactory, save it by clicking on
![]() .
.
6.3.2 Default parameters
Used function and default parameters:
If a variable has missing values but is constant otherwise, the missing values are filled with the unique value.
Then, according to the type of imputation:
Method zeros:
- missing values are fille with 0
Method PCA:
If the variables to impute are numeric:
missMDA::imputePCA(df, ncp=2, scale = FALSE)If the variables to impute are categoricals:
missMDA::imputeMCA(df, ncp=2)If the variables to impute are mixed:
missMDA::imputeFAMD(df, ncp=2)
Method KNN:
VIM::kNN(df, imp_var = FALSE)